library(tidyverse)
library(fuzzyjoin)
library(gt)
fair_use_cases <- readr::read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2023/2023-08-29/fair_use_cases.csv')
fair_use_findings <- readr::read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2023/2023-08-29/fair_use_findings.csv')From the dataset description:
The data this week comes from the U.S. Copyright Office Fair Use Index.
Fair use is a longstanding and vital aspect of American copyright law. The goal of the Index is to make the principles and application of fair use more accessible and understandable to the public by presenting a searchable database of court opinions, including by category and type of use (e.g., music, internet/digitization, parody).
There are two datasets this week for which the rows align, but the values might not precisely line up for a clean join – a case you often have to deal with in real-world data.
This last point is what I’ll be focusing on in this post: The challenge of joining two datasets together that don’t line up for a clean join.
Glimpse
glimpse(fair_use_cases)Rows: 251
Columns: 7
$ case <chr> "De Fontbrune v. Wofsy, 39 F.4th 1214 (9th Cir. 2022)",…
$ year <dbl> 2022, 2022, 2022, 2022, 2022, 2022, 2022, 2022, 2022, 2…
$ court <chr> "9th Circuit", "C.D. Cal.", "S.D.N.Y.", "D.D.C.", "2d C…
$ jurisdiction <chr> "9th Circuit", "9th Circuit", "2nd Circuit", "District …
$ categories <chr> "Education/Scholarship/Research; Photograph", "Painting…
$ outcome <chr> "Fair use not found", "Preliminary finding; Fair use no…
$ fair_use_found <lgl> FALSE, FALSE, TRUE, FALSE, TRUE, FALSE, FALSE, TRUE, TR…glimpse(fair_use_findings)Rows: 251
Columns: 9
$ title <chr> "De Fontbrune v. Wofsy", "Sedlik v. Von Drachenberg", "Ske…
$ case_number <chr> "39 F.4th 1214 (9th Cir. 2022)", "No. CV 21-1102, 2022 WL …
$ year <chr> "2022", "2022", "2022", "2022", "2022", "2022", "2022", "2…
$ court <chr> "United States Court of Appeals for the Ninth Circuit", "U…
$ key_facts <chr> "Plaintiffs own the rights to a catalogue comprised of 16,…
$ issue <chr> "Whether reproduction of photographs documenting artwork i…
$ holding <chr> "The panel held that the first factor, the purpose and cha…
$ tags <chr> "Education/Scholarship/Research; Photograph", "Painting/Dr…
$ outcome <chr> "Fair use not found", "Preliminary finding; Fair use not f…Approach 0: column binding
I’ll start with the simplest approach: Could a bind_cols() work?
No, there is mis-alignment as we can see in the table of results: case and title do not match.
bind_cols_join %>%
select(case, title) %>%
head(111) %>%
tail(3) %>%
gt() %>%
opt_interactive() %>%
tab_header(title = "bind_cols() results") %>%
tab_options(table.background.color = '#f1f3f5',
ihtml.page_size_default = 3)Approach 1: Fuzzy joining
I noticed that title tends to be a substring of case. Given this, I can try “fuzzy joining” using the fuzzyjoin library. I’ll also join with year, just in case there are multiple substring matches this should help to better resolve them to their correct match.
fair_use_cases %>%
fuzzy_left_join(fair_use_findings %>% select(-court, -outcome),
by = c("case" = "title", "year" = "year"),
match_fun = str_detect) %>%
select(-year.y) %>%
rename(year = year.x) -> fuzzy_join
(fuzzy_join %>%
filter(is.na(title)) %>%
distinct(case, title) %>%
count())$n[1] 37This seems promising: only 37 records did not match. Can I fine-tune it?
I find that it always helps to look at examples of errors and see what can be learned.
## Identify the missing cases from each set
fuzzy_join %>%
filter(is.na(title)) %>%
select(colnames(fair_use_cases)) -> cases_without_match
fair_use_findings %>%
filter(!title %in% fuzzy_join$title) -> findings_without_match
cases_without_match %>%
select(case, year) %>%
arrange(case, year) %>%
gt() %>%
opt_interactive() %>%
tab_header(title = "Cases without a match in Findings") %>%
tab_options(table.background.color = '#f1f3f5',
ihtml.page_size_default = 3)findings_without_match %>%
select(title, case_number, year) %>%
arrange(title, year) %>%
gt() %>%
opt_interactive() %>%
tab_header(title = "Findings without a match in Cases") %>%
tab_options(table.background.color = '#f1f3f5',
ihtml.page_size_default = 3)Some observations about these examples that may have caused mis-matching:
Non-ASCII characters like
óor ```Serv.vs.Servs.Punctuation inconsistencies, like
Nat'l Acad. of TV Arts & Scis., Inc.vs.Nat'l Acad. of TV Arts & Scis., Inc.,LLCis sometimes missing, likeTresona Multimedia, LLCvs.Tresona Multimedia v.
These steps could improve the match rate:
Convert to ASCII
Convert to lowercase
Remove occurrences of
LLC
I’ll perform these standardizations and try joining again.
Approach 2: Fuzzy joining + string cleaning
# Mutate a standardized variable: case_std
fair_use_cases %>%
mutate(case_std = tolower(case),
case_std = iconv(case_std, to='ASCII//TRANSLIT'),
case_std = str_replace_all(case_std, "[^[:alnum:][:space:]]", ""),
case_std = str_replace_all(case_std, 'llc', '')) -> fair_use_cases_std
# Mutate a standardized variable: title_std
fair_use_findings %>%
mutate(year = str_sub(year, 1, 4),
title_std = tolower(title),
title_std = iconv(title_std, to='ASCII//TRANSLIT'),
title_std = str_replace_all(title_std, "[^[:alnum:][:space:]]", ""),
title_std = str_replace_all(title_std, 'llc', '')) -> fair_use_findings_std
# Perform a fuzzy join
fair_use_cases_std %>%
fuzzy_left_join(fair_use_findings_std %>% select(-year, -court, -outcome),
by = c("case_std" = "title_std"),
match_fun = str_detect) -> fuzzy_join
## Identify the missing cases from each set
fuzzy_join %>%
filter(is.na(title)) %>%
select(colnames(fair_use_cases)) -> fair_use_cases_remainder
fair_use_findings_std %>%
filter(!title %in% fuzzy_join$title) -> fair_use_findings_remainder
fair_use_cases_remainder %>%
select(case, year) %>%
arrange(case, year) %>%
gt() %>%
opt_interactive() %>%
tab_header(title = "Cases without a match in Findings") %>%
tab_options(table.background.color = '#f1f3f5',
ihtml.page_size_default = 3)fair_use_findings_remainder %>%
select(title, case_number, year) %>%
arrange(title, year) %>%
gt() %>%
opt_interactive() %>%
tab_header(title = "Findings without a match in Cases") %>%
tab_options(table.background.color = '#f1f3f5',
ihtml.page_size_default = 3)I can see that 16 are missing in each case, and what’s interesting about these two sets is they appear to be the same items, in the same order. This means we can simply do a bind_cols() operation to join them together.
Final solution: Fuzzy joining + string cleaning + column binding
Here I put all of the lessons learned so far in fuzzy joining together into one final chunk of code.
# Mutate a standardized variable: case_std
fair_use_cases %>%
mutate(case_std = tolower(case),
case_std = iconv(case_std, to='ASCII//TRANSLIT'),
case_std = str_replace_all(case_std, "[^[:alnum:][:space:]]", ""),
case_std = str_replace_all(case_std, 'llc', '')) %>%
select(-court, -outcome) -> fair_use_cases_std
# Mutate a standardized variable: title_std
fair_use_findings %>%
mutate(year = str_sub(year, 1, 4),
title_std = tolower(title),
title_std = iconv(title_std, to='ASCII//TRANSLIT'),
title_std = str_replace_all(title_std, "[^[:alnum:][:space:]]", ""),
title_std = str_replace_all(title_std, 'llc', '')) -> fair_use_findings_std
# Perform a fuzzy join
fair_use_cases_std %>%
fuzzy_left_join(fair_use_findings_std,
by = c("case_std" = "title_std", "year" = "year"),
match_fun = str_detect) %>%
select(-case_std,
-title_std,
-year.y) %>%
rename(year = year.x) -> fuzzy_join
# Identify the missing cases from each set
fuzzy_join %>%
filter(is.na(title)) %>%
select(colnames(fair_use_cases)) -> fair_use_cases_remainder
fair_use_findings_std %>%
select(-year, -court, -outcome) %>%
filter(!title %in% fuzzy_join$title) -> fair_use_findings_remainder
# Bind the columns together
fair_use_cases_remainder %>%
select(colnames(fair_use_cases)) %>%
bind_cols(fair_use_findings_remainder) -> remainders_join
# Join together the two subsets
fuzzy_join %>%
filter(!is.na(title)) %>%
bind_rows(remainders_join) %>%
arrange(case, title, year) -> final_joinValidation checks
When inspecting the final result I expect to see 251 rows; none of which should be duplicates, and I do.
final_join %>%
summarize(
n_rows = n(),
n_distinct_rows = n_distinct(case, title)
)# A tibble: 1 × 2
n_rows n_distinct_rows
<int> <int>
1 251 251As another check, I can look at the same records that were misaligned with the first approach. I can see they are now aligned.
final_join %>%
select(case, title, case_number) %>%
head(111) %>%
tail(3) %>%
gt() %>%
opt_interactive() %>%
tab_header(title = "Rows that were previously misaligned with bind_cols() are now aligned") %>%
tab_options(table.background.color = '#f1f3f5',
ihtml.page_size_default = 3)And here is the full table.
final_join %>%
select(case, title, case_number) %>%
gt() %>%
opt_interactive() %>%
tab_header(title = "Final results table (case, title, case_number)") %>%
tab_options(table.background.color = '#f1f3f5',
ihtml.page_size_default = 3)It looks like the matches are now correct.
Data visualization
And now for a quick data viz showing the journey towards achieving 100% matches between the two data frames.
data.frame("method" = c("fuzzy joining",
"fuzzy joining + \nstring cleaning",
"fuzzy joining + \nstring cleaning + \ncolumn binding"),
"match_rate" = c((251-37)/251*100, (251-16)/251*100, 100),
"class" = c(FALSE, FALSE, TRUE)) %>%
ggplot(aes(x = method, y = match_rate, fill=class)) +
geom_bar(stat="identity", colour="black") +
geom_text(aes(label = paste0(round(match_rate), "%")), colour="black", vjust=-.2) +
xlab("Method") +
ylab("Match Rate") +
ggtitle("Percentage of matches found between<br><span style='font-family:monospace;color:red'>fair_use_cases</span> and <span style='font-family:monospace;color:red'>fair_use_findings</span><br>by method(s) used") +
ggthemes::theme_fivethirtyeight() +
gghighlight::gghighlight(class == TRUE, unhighlighted_params = list(fill="grey", colour="black")) +
theme(plot.title = ggtext::element_markdown(),
plot.caption = ggtext::element_markdown())label_key: method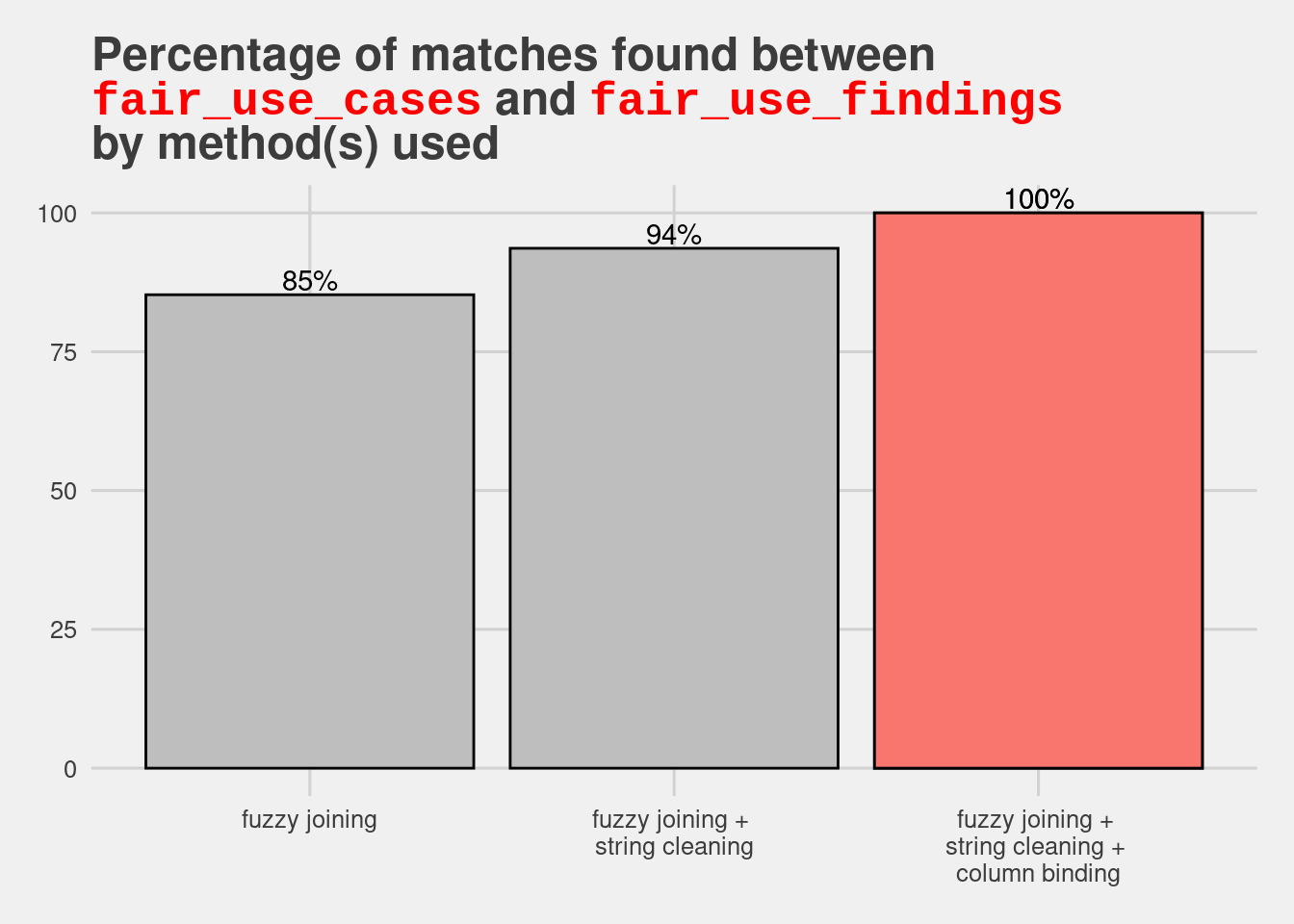
ggsave('social-image.png', height=5, width=10)